Data Frames in Python#
A data frame is a two-dimensional, size-mutable, and heterogeneous data structure. Think of it as a table or spreadsheet where rows represent records and columns represent variables or attributes. In Python, the pandas.DataFrame class is used to represent data frames.
Key characteristics of data frames:
Columns can contain different data types (e.g., numeric, character, factor).
Rows represent individual observations or cases.
Data frames can hold large datasets efficiently.
Creating Data Frames#
A data frame can be created from various data types:
import pandas as pd
# Creating a data from a dictionary
data = {
"Name": ["Amrit", "Ishwor", "Indra", "Rakshya"],
"Age": [27, 24, 21, 25],
"Grade": ["A", "B", "A", "B"],
"Major": ["Maths", "Science", "Maths", "Science"]
}
df = pd.DataFrame(data)
# Display the data frame
print(df)
Name Age Grade Major
0 Amrit 27 A Maths
1 Ishwor 24 B Science
2 Indra 21 A Maths
3 Rakshya 25 B Science
Accessing and Modifying Data#
Selecting Rows and Columns
We can access elements of a data frame using indexing, column names, or logical conditions.
# Access a column
print(df["Name"])
0 Amrit
1 Ishwor
2 Indra
3 Rakshya
Name: Name, dtype: object
# Access a row by index
print(df.loc[0])
Name Amrit
Age 27
Grade A
Major Maths
Name: 0, dtype: object
# Access a specific value
value = df.at[2, "Name"] # Using label-based indexing
print(value)
Indra
# Using position-based indexing
value = df.iloc[2, 0]
print(value)
Indra
Adding and Removing Columns
We can add a new column to a data frame dynamically or remove an existing column.
# Add a column 'Pass' where students with Grade 'A' are True
df["Pass"] = df["Grade"] == "A"
print(df)
Name Age Grade Major Pass
0 Amrit 27 A Maths True
1 Ishwor 24 B Science False
2 Indra 21 A Maths True
3 Rakshya 25 B Science False
# Remove the 'Grade' column
df = df.drop(columns=["Grade"])
print(df)
Name Age Major Pass
0 Amrit 27 Maths True
1 Ishwor 24 Science False
2 Indra 21 Maths True
3 Rakshya 25 Science False
Data Frame Operations#
Filtering and Subsetting
We can filter rows based on conditions using logical operators.
# Filter rows where Age > 22
filtered_df = df[df["Age"] > 22]
print(filtered_df)
Name Age Major Pass
0 Amrit 27 Maths True
1 Ishwor 24 Science False
3 Rakshya 25 Science False
Sorting and Reordering
Sorting a data frame is straightforward with the sort_values() method.
# Sort by the 'Age' column in ascending order
df_sorted = df.sort_values(by="Age", ascending=True)
print(df_sorted)
Name Age Major Pass
2 Indra 21 Maths True
1 Ishwor 24 Science False
3 Rakshya 25 Science False
0 Amrit 27 Maths True
Summary and Statistical Functions
The .describe() method in pandas provides a summary of each column in a DataFrame. Here’s how:
# Summary of the DataFrame (Descriptive statistics)
print(df.describe())
Age
count 4.00
mean 24.25
std 2.50
min 21.00
25% 23.25
50% 24.50
75% 25.50
max 27.00
# Get number of rows and columns
num_rows, num_cols = df.shape
print(f"Number of rows: {num_rows}")
print(f"Number of columns: {num_cols}")
Number of rows: 4
Number of columns: 4
Aggregation and Grouping
The groupby method in pandas along with aggregation functions enable aggregation and grouping.
# Group by 'Major' and calculate the average age
grouped_df = df.groupby("Major").agg(Average_Age=("Age", "mean")).reset_index()
print(grouped_df)
Major Average_Age
0 Maths 24.0
1 Science 24.5
groupbygroups the data frame by the ‘Major’ columnaggapplies the mean function to the ‘Age’ column and renames the result to ‘Average_Age’reset_index()resets the index so the grouped column “Major” becomes a regular column in the resulting data frame.
So far, this chapter has introduced you to the versatility and power of data frames in Python, laying the foundation for data manipulation and analysis. Now, we will delve into data frames specifically designed for geospatial data.
Geospatial Data Frames#
A Geospatial Data Frame (GeoDataFrame) extends a standard data frame by incorporating a geometry column that holds spatial data, such as points, lines, and polygons. This combination of spatial and attribute data makes geospatial data frames particularly useful for spatial analysis and mapping.
In Python, the GeoPandas library is often used to handle geospatial data frames, providing easy-to-use structures for working with vector data and offering compatibility with other libraries for geospatial and statistical analysis.
The geometry is a key component of geospatial data frames, defining the spatial features for each observation. Each row represents a spatial feature linked to its attributes, and the geometry column typically stores spatial data in formats such as GeoJSON, WKT (Well-Known Text), or binary formats like WKB (Well-Known Binary).
Creating GDF#
You can create a geospatial data frame using GeoPandas. Here’s an example:
import geopandas as gpd
import pandas as pd
from shapely.geometry import Point
# Create a data frame with spatial data
data = {
"Name": ["Point A", "Point B", "Point C"],
"Latitude": [27.7, 28.2, 26.9],
"Longitude": [85.3, 84.9, 83.8]
}
df = pd.DataFrame(data)
# Convert to a GeoDataFrame
geometry = [Point(xy) for xy in zip(df["Longitude"], df["Latitude"])]
geo_data = gpd.GeoDataFrame(df, geometry=geometry)
# Set the coordinate reference system (CRS) to WGS 84
geo_data.set_crs(epsg=4326, inplace=True)
# Print the GeoDataFrame
print(geo_data)
Name Latitude Longitude geometry
0 Point A 27.7 85.3 POINT (85.3 27.7)
1 Point B 28.2 84.9 POINT (84.9 28.2)
2 Point C 26.9 83.8 POINT (83.8 26.9)
Manipulating GDF#
Spatial Queries
Geospatial data frames support operations like spatial filtering and proximity analysis.
from shapely.geometry import box
# Define a bounding box and convert it to a geometry object
bbox = box(84, 27, 86, 29) # xmin, ymin, xmax, ymax
bbox_polygon = gpd.GeoSeries([bbox], crs="EPSG:4326")
# Filter points within the bounding box
filtered_data = geo_data[geo_data.geometry.within(bbox_polygon.iloc[0])]
# Print filtered data
print(filtered_data)
Name Latitude Longitude geometry
0 Point A 27.7 85.3 POINT (85.3 27.7)
1 Point B 28.2 84.9 POINT (84.9 28.2)
Attribute-Based Operations
Just like standard data frames, we can manipulate attributes in geospatial data frames.
# Add a new column for Elevation
geo_data['Elevation'] = [1500, 2000, 1700]
# Print the updated GeoDataFrame
print(geo_data)
Name Latitude Longitude geometry Elevation
0 Point A 27.7 85.3 POINT (85.3 27.7) 1500
1 Point B 28.2 84.9 POINT (84.9 28.2) 2000
2 Point C 26.9 83.8 POINT (83.8 26.9) 1700
Visualization of GDF#
Geospatial data frames can be visualized using the matplotlib library.
import matplotlib.pyplot as plt
# Plot the GeoDataFrame
geo_data.plot(marker='o', color='blue', legend=True)
plt.title("Geospatial Data Points")
plt.show()

Geospatial data frames bridge the gap between tabular data and spatial analysis, making them indispensable for modern geospatial workflows. Their versatility, combined with Python’s ecosystem of geospatial libraries, provides an efficient and intuitive way to analyze and visualize spatial data.
Vector GeoDataFrame#
In Python, the GeoDataFrame from the GeoPandas library is used to represent vector-based spatial data. A GeoDataFrame combines both spatial and tabular data, making it ideal for spatial analysis and data manipulation. It is essentially a data frame with a special geometry column that contains spatial data, such as points, lines, and polygons.
With a GeoDataFrame, you can perform typical data manipulation tasks, such as filtering, summarizing, and joining, on the tabular data. At the same time, it allows for spatial operations like plotting, spatial indexing, and spatial joins. The tabular portion of a GeoDataFrame allows you to perform standard data frame operations (like groupby, sort, merge), while the geometry column enables you to carry out spatial analysis and visualization.
This section will focus on working with geospatial data, such as the vector data for the Kanchanpur district in Nepal, available in GeoPackage (.gpkg) format. You can download this data, load it using GeoPandas, and perform both spatial and tabular operations. The GeoDataFrame provides a powerful tool for handling, analyzing, and visualizing geographic data, seamlessly integrating both spatial and non-spatial aspects of analysis.
Structure#
The vector GeoDataFrame contains both the geometry column and attribute data.
import geopandas as gpd
# Import the GeoDataFrame
vector_data = gpd.read_file("data/vector/kanchanpur.gpkg")
# View the structure of the GeoDataFrame
print(vector_data.info())
<class 'geopandas.geodataframe.GeoDataFrame'>
RangeIndex: 21 entries, 0 to 20
Data columns (total 2 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 NAME 21 non-null object
1 geometry 21 non-null geometry
dtypes: geometry(1), object(1)
memory usage: 464.0+ bytes
None
# Display the first few rows
print(vector_data.head())
NAME geometry
0 BaisiBichawa MULTIPOLYGON (((80.49934 28.64667, 80.49709 28...
1 Beldandi MULTIPOLYGON (((80.25229 28.75782, 80.25377 28...
2 Chandani MULTIPOLYGON (((80.10973 28.98432, 80.10986 28...
3 Daijee MULTIPOLYGON (((80.34424 29.05416, 80.34449 29...
4 Dekhatbhuli MULTIPOLYGON (((80.44701 28.78921, 80.43608 28...
Adding New Columns#
In Python, you can add a new column to a GeoDataFrame in a similar way you would add it to a regular DataFrame using pandas.
# Add a new column - Population
vector_data['Population'] = [
37900, 46565, 72813, 49223, 52221, 64436, 40823,
97216, 36368, 277108, 51835, 38199, 46773, 44571,
48838, 11581, 131, 19211, 56755, 58257, 37573
]
# Display the updated GeoDataFrame
print(vector_data.head())
NAME geometry Population
0 BaisiBichawa MULTIPOLYGON (((80.49934 28.64667, 80.49709 28... 37900
1 Beldandi MULTIPOLYGON (((80.25229 28.75782, 80.25377 28... 46565
2 Chandani MULTIPOLYGON (((80.10973 28.98432, 80.10986 28... 72813
3 Daijee MULTIPOLYGON (((80.34424 29.05416, 80.34449 29... 49223
4 Dekhatbhuli MULTIPOLYGON (((80.44701 28.78921, 80.43608 28... 52221
Arithmetic Computation#
In Python, you can calculate the area of each geometry in a GeoDataFrame and create a new column for population density as follows:
# Reproject vector data to a projected CRS (e.g., UTM zone 45N for this area)
vector_data = vector_data.to_crs(epsg=32645)
# Calculate the area of each district (in square meters)
vector_data['area_m2'] = vector_data.geometry.area
# Convert the area from square meters to square kilometers
vector_data['area_km2'] = vector_data['area_m2'] / 1e6
# Add a new column with density (population/area)
vector_data['Density'] = vector_data['Population'] / vector_data['area_km2']
# Display the updated GeoDataFrame
print(vector_data[['NAME', 'Population', 'area_km2', 'Density']].head())
NAME Population area_km2 Density
0 BaisiBichawa 37900 74.476642 508.884384
1 Beldandi 46565 24.906232 1869.612393
2 Chandani 72813 32.517201 2239.214842
3 Daijee 49223 102.823123 478.715280
4 Dekhatbhuli 52221 91.109522 573.167312
Selecting and Filtering#
You can filter the data using GeoDataFrame from geopandas. Here’s how you can select specific columns and display the first few rows:
# Select specific columns (e.g., "NAME" and "Population")
selected_data = vector_data[['NAME', 'Population']]
# Display the first few rows
print(selected_data.head())
NAME Population
0 BaisiBichawa 37900
1 Beldandi 46565
2 Chandani 72813
3 Daijee 49223
4 Dekhatbhuli 52221
# Filter rows where Population is greater than 60,000
filtered_data = vector_data[vector_data['Population'] > 60000]
# Display the filtered data (similar to tibble in R)
print(filtered_data[['NAME', 'Population', 'Density', 'area_km2']])
NAME Population Density area_km2
2 Chandani 72813 2239.214842 32.517201
5 Dodhara 64436 2591.163947 24.867589
7 Kalika 97216 3789.177761 25.656226
9 Laxmipur 277108 9518.765708 29.111758
Grouping and Summarizing#
Grouping and summarizing operations are common in data analysis. You can group data based on an attribute and calculate summary statistics for each group.
# Group by 'NAME' and calculate the total population and average density
region_summary = vector_data.groupby('NAME').agg(
total_population=('Population', 'sum'),
average_density=('Density', 'mean')
).reset_index()
# Display the result
print(region_summary)
NAME total_population average_density
0 BaisiBichawa 37900 508.884384
1 Beldandi 46565 1869.612393
2 Chandani 72813 2239.214842
3 Daijee 49223 478.715280
4 Dekhatbhuli 52221 573.167312
5 Dodhara 64436 2591.163947
6 Jhalari 40823 332.137920
7 Kalika 97216 3789.177761
8 Krishnapur 36368 218.213683
9 Laxmipur 277108 9518.765708
10 MahendranagarN.P. 51835 299.845566
11 Parasan 38199 1004.067157
12 Pipaladi 46773 1087.800781
13 RaikawarBichawa 44571 545.906632
14 RampurBilaspur 48838 1236.669117
15 RauteliBichawa 11581 382.064494
16 Royal Shuklaphanta 131 0.360413
17 Shankarpur 19211 538.994951
18 Sreepur 56755 1012.202386
19 Suda 58257 1001.135055
20 Tribhuwanbasti 37573 1911.038295
groupby('NAME'): Groups the GeoDataFrame by the NAME column, which represents different villages in Kanchanpur district.agg(): Used to specify the aggregation functions. Here, it calculates the sum ofPopulationand the mean ofDensity.reset_index(): Resets the index after the grouping operation to make sure the resultingDataFrameis formatted properly.
Pivoting and Reshaping#
In Python, you can achieve pivoting and reshaping using pandas.
# Drop geometry column and pivot data into long format
long_format = vector_data.drop(columns='geometry') \
.melt(id_vars=["NAME"], value_vars=["Population", "Density"],
var_name="attribute", value_name="value")
# Display the first and middle few rows of the reshaped DataFrame
print(long_format.iloc[[0,1,2,21,22,23], :])
NAME attribute value
0 BaisiBichawa Population 37900.000000
1 Beldandi Population 46565.000000
2 Chandani Population 72813.000000
21 BaisiBichawa Density 508.884384
22 Beldandi Density 1869.612393
23 Chandani Density 2239.214842
drop(columns='geometry'): This removes the geometry column since it’s not needed for reshaping the tabular data.melt(): This function is used to pivot data from wide to long format.
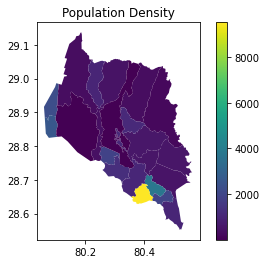
Visualization#
To plot the population density from a GeoDataFrame using matplotlib and geopandas, you can do the following:
import geopandas as gpd
import matplotlib.pyplot as plt
# Reproject vector data to a projected CRS (e.g., UTM zone 45N for this area)
vector_data = vector_data.to_crs(epsg=4326)
# Plot the Population Density
vector_data.plot(column='Density', cmap='viridis', legend=True)
plt.title("Population Density")
plt.show()
Raster GeoDataFrame#
A Raster GeoDataFrame extends the concept of a standard GeoDataFrame to represent raster data in a tabular format, integrating spatial information with associated metadata. While raster data is inherently grid-based, a Raster GeoDataFrame provides a structured way to store additional attributes, such as statistics, classification information, or metadata, alongside spatial references. This format is particularly useful for working with tiled or multi-resolution raster datasets, enabling seamless integration with vector data workflows. By combining the tabular nature of data frames with the grid structure of raster data, it facilitates efficient querying, visualization, and analysis of geospatial raster datasets.
Creating a Raster Data#
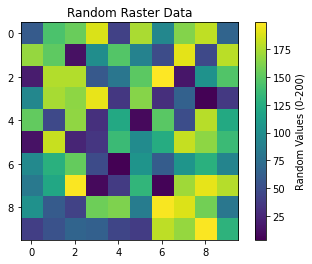
The python uses numpy to create random raster data and matplotlib to visualize it. The rasterio package is included to define the raster’s metadata and coordinate reference, though it isn’t required for basic visualization. If you’d like, this data can be saved to a file using rasterio.
Creating a raster dataset with random values and plotting it:
import numpy as np
import rasterio
from rasterio.plot import show
import matplotlib.pyplot as plt
# Create a raster-like array with random values
rows, cols = 10, 10
raster_data = np.random.uniform(0, 200, (rows, cols))
# Define metadata for the raster
transform = rasterio.transform.from_origin(west=0, north=10, xsize=1, ysize=1) # Example transform
meta = {
'driver': 'GTiff',
'dtype': 'float32',
'nodata': None,
'width': cols,
'height': rows,
'count': 1,
'crs': '+proj=latlong',
'transform': transform
}
# Plot the raster
plt.imshow(raster_data, cmap='viridis')
plt.colorbar(label="Random Values (0-200)")
plt.title("Random Raster Data")
plt.show()
Coverting Raster to GDF#
Here’s how you can convert raster data to a DataFrame in Python and view the first few rows/columns:
import pandas as pd
import numpy as np
# Convert the raster array to a DataFrame
df = pd.DataFrame(raster_data)
# Display the first few rows
print(df.iloc[[0, 1, 2, 3], [0, 1, 2, 3]])
0 1 2 3
0 59.735910 144.102702 154.567396 188.186372
1 168.309111 150.893560 12.511152 99.293095
2 18.774834 176.693506 176.326299 58.348779
3 94.479577 173.212253 166.123110 192.648874
If you’d like to include spatial information (e.g., row and column indices as spatial coordinates), here’s how you can do it:
# Add spatial coordinates (row and column indices)
x, y = np.meshgrid(range(cols), range(rows)) # Create grid of coordinates
df_with_coords = pd.DataFrame({
"x": x.flatten(),
"y": y.flatten(),
"value": raster_data.flatten()
})
# Display the first few rows
print(df_with_coords.head())
x y value
0 0 0 59.735910
1 1 0 144.102702
2 2 0 154.567396
3 3 0 188.186372
4 4 0 41.736887
This approach provides both the raster values and their respective spatial coordinates, enabling easier manipulation and analysis of the data.
Summarize Raster Values#
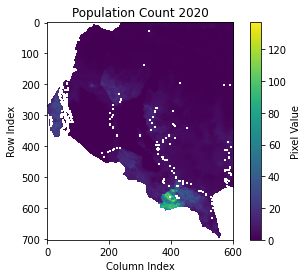
In this subsection, we will load the Population Count raster data for the year 2020.
import rasterio
import matplotlib.pyplot as plt
# Load the raster data
raster_path = "data/raster/popCount_2020.tif"
with rasterio.open(raster_path) as src:
raster_data = src.read(1)
# Plot the raster using Matplotlib
plt.imshow(raster_data, cmap='viridis')
plt.colorbar(label="Pixel Value")
plt.title("Population Count 2020")
plt.xlabel("Column Index")
plt.ylabel("Row Index")
plt.show()
We can calculate basic summary statistics (mean, median, min, max) to get an overview of the population distribution across the region.
import numpy as np
raster_file = "data/raster/popCount_2020.tif"
# open the raster file
with rasterio.open(raster_file) as src:
# Read the raster data as a numpy array
raster_data = src.read(1) # Single band raster
# Mask invalid or nodata values
# Define a custom condition for masking (e.g., mask zeros)
masked_data = np.ma.masked_where((raster_data == 0) | np.isnan(raster_data), raster_data)
# Calculate summary statistics
min_value = masked_data.min()
max_value = masked_data.max()
mean_value = masked_data.mean()
median_value = np.ma.median(masked_data)
# Print the results
print(f"Minimum value: {min_value}")
print(f"Maximum value: {max_value}")
print(f"Mean value: {mean_value}")
print(f"Median value: {median_value}")
Minimum value: 0.1460667997598648
Maximum value: 136.91754150390625
Mean value: 5.2824760935231385
Median value: 2.4147253036499023
Identify High-Density Regions#
You can extract specific regions of the raster where the population is above a certain threshold, which could be useful for identifying high-density areas.
import rasterio
import numpy as np
raster_file = "data/raster/popCount_2020.tif"
# Define the population density threshold
threshold = 100
with rasterio.open(raster_file) as src:
raster_data = src.read(1) # Read the first band
transform = src.transform # Get the affine transform
# Mask invalid or nodata values
# Define a custom condition for masking (e.g., mask zeros)
masked_data = np.ma.masked_where((raster_data == 0) | np.isnan(raster_data), raster_data)
# Identify high-density cells
high_density_mask = raster_data > threshold
high_density_indices = np.argwhere(high_density_mask)
# Convert indices to geographic coordinates
coordinates_high_density = [
rasterio.transform.xy(transform, row, col, offset="center")
for row, col in high_density_indices
]
num_high_density_cells = len(coordinates_high_density)
print(f"Number of high-density cells: {num_high_density_cells}")
print(f"Sample coordinates of high-density cells: {coordinates_high_density[:3]}") # Display first three coordinates
Number of high-density cells: 549
Sample coordinates of high-density cells: [(80.399999692985, 28.688333420150848), (80.39583302633488, 28.685833420161014), (80.3966663596649, 28.685833420161014)]
import matplotlib.pyplot as plt
# Coordinates of high-density cells (xyFromCell provides (x, y) coordinates)
x_coords, y_coords = zip(*coordinates_high_density)
# Plot the high-density points
plt.figure(figsize=(8, 6))
plt.scatter(x_coords, y_coords, c='red', s=5, label="High Density Cells")
plt.title("High Density Regions")
plt.xlabel("Longitude")
plt.ylabel("Latitude")
plt.legend()
plt.show()
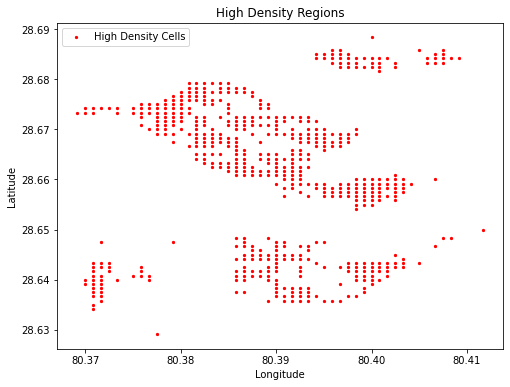
x_coords, y_coords: Unzips the coordinates_high_density to separate x and y values.scatter: Plots the high-density cells as red points on the plot.s=5: Sets the size of the points.
Classify Raster Data#
To classify raster data into categories (e.g., low, medium, high) in Python, you can use numpy’s cut function for creating intervals.
import numpy as np
import pandas as pd
import rasterio
# Open the raster data
with rasterio.open("data/raster/popCount_2020.tif") as src:
raster_data = src.read(1) # Read the first band (assuming single-band raster)
# Reclassify the raster values into categories
breaks = [0, 1, 5, 20, 100, np.inf] # Define the class boundaries
labels = ["Very Low", "Low", "Medium", "High", "Very High"]
# Use np.digitize to classify the raster values
classified_data = np.digitize(raster_data, bins=breaks, right=True)
# Handle the edge case where np.digitize assigns a value greater than the length of the labels
classified_data[classified_data == len(labels) + 1] = len(labels) # Ensure index for 'Very High'
# Convert the numeric classification to labels
classified_labels = np.array(labels)[classified_data - 1] # Adjust index for 0-based indexing
# Create a DataFrame with x, y coordinates and classification
rows, cols = np.indices(raster_data.shape)
df_classified = pd.DataFrame({
'x': cols.flatten(),
'y': rows.flatten(),
'class': classified_labels.flatten()
})
# Show the first few rows of the classified DataFrame
print(df_classified.head())
x y class
0 0 0 Very High
1 1 0 Very High
2 2 0 Very High
3 3 0 Very High
4 4 0 Very High
np.digitize: Classifies the raster values based on predefinedbreaks. The function returns the indices of which class each value belongs to.classified_labels: Uses the indices fromnp.digitizeto assign the corresponding labels from thelabelslist.Creating DataFrame: The coordinates of each raster cell(x, y)are flattened into arrays and combined with the classified labels to create aDataFrame.right=True: Ensures that the right boundary of each bin is included in the classification. For example, values exactly equal to 5 will be classified as “Low”, rather than being excluded.Edge Case Handling: If np.digitize assigns an index greater than the size of labels (for values above the last bin), we correct it by explicitly setting those values to the last class (“Very High”).Adjustment for 0-based Indexing: Since the labels array is 0-based, we subtract 1 from the classification indices to correctly match the class labels.
This code will provide the classified raster values as a DataFrame, which you can use for further analysis or visualization.
from matplotlib.colors import ListedColormap
# Create a plot to visualize the classified data
fig, ax = plt.subplots(figsize=(8, 6))
# Custom colors for the classes
cmap = ListedColormap(['#f9f9f9', '#e1f7d5', '#d4f1a1', '#e7d300', '#ff6347'])
bounds = [0, 1, 5, 20, 100, np.inf]
norm = plt.Normalize(vmin=0, vmax=len(labels)-1)
# Plot the classified raster data
cax = ax.imshow(classified_data, cmap=cmap, norm=norm)
ax.set_title("Classified Population Density")
# Add a colorbar with the class labels
colorbar = fig.colorbar(cax, ax=ax, ticks=np.arange(len(labels)))
colorbar.set_ticklabels(labels)
colorbar.set_label('Classification')
# Show the plot
plt.show()
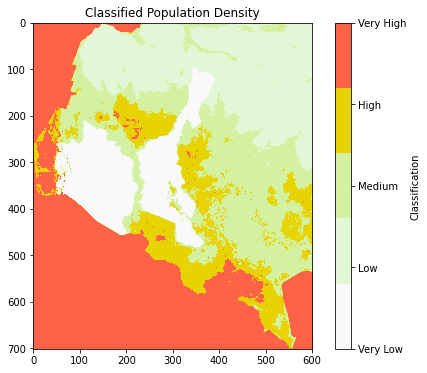
Note: Values outside of your Area of Interest (AOI) show as “Very High,” likely occurs because the raster data outside the AOI is being treated as having the highest value by default. To solve this, you need to mask the raster to focus only on the AOI, ensuring that areas outside the AOI are not processed or classified.
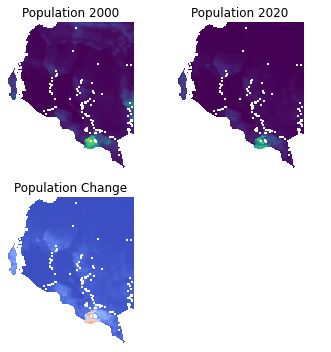
Change Identification#
If you have multiple rasters representing population data over different time periods, you can compute the changes in population between two periods. Let’s load the Population Count raster data for years 2000 and 2020, so we can compute the changes in population over a 20-year period.
import rasterio
import numpy as np
import matplotlib.pyplot as plt
# Load population count raster for the year 2000
r1 = rasterio.open("data/raster/popCount_2000.tif")
# Load population count raster for the year 2020
r2 = rasterio.open("data/raster/popCount_2020.tif")
# Read the population count data as arrays
pop_2000 = r1.read(1)
pop_2020 = r2.read(1)
# Population change between 2020 and 2000
population_change = pop_2020 - pop_2000
# Set up a plotting layout: 2 rows, 2 columns
fig, axs = plt.subplots(2, 2, figsize=(5, 5))
# Plot the raster using Matplotlib
axs[0, 0].imshow(pop_2000, cmap='viridis')
axs[0, 0].set_title("Population 2000")
axs[0, 0].axis('off')
# Plot the raster using Matplotlib
axs[0, 1].imshow(pop_2020, cmap='viridis')
axs[0, 1].set_title("Population 2020")
axs[0, 1].axis('off')
# Plot the raster using Matplotlib
axs[1, 0].imshow(population_change, cmap='coolwarm')
axs[1, 0].set_title("Population Change")
axs[1, 0].axis('off')
# Reset the layout for default (1 plot)
axs[1, 1].axis('off') # Empty subplot as we have only 3 plots
plt.tight_layout()
plt.show()
Reading the Raster Files: – The
rasterio.open()function is used to open the raster files. –r1.read(1)andr2.read(1)read the first band (the population count data) of the raster files as arrays.Population Change Calculation: – The population change is calculated by subtracting the 2000 population values (
pop_2000) from the 2020 population values (pop_2020).Plotting: –
matplotlibis used to create the plots. We useimshowto display the rasters with theviridiscolormap for population count andcoolwarmfor population change. – The layout is set withplt.subplots()to create 2 rows and 2 columns, similar topar(mfrow = c(2, 2))in R. – We turn off the axes withaxis('off')for a cleaner plot.
For this tutorial, this concludes the coverage of Data Frames in Python. If you would like to explore additional examples, or need clarification on any of the steps covered, please visit the GitHub repository: Python_tutorial and feel free to open an issue.
